About Me
I’m a machine learning researcher interested in representation learning (generative and self-supervised) and interpretability of machine learning models.
I did my PhD advised by Ke Yuan at the University of Glasgow, Computing Science department. The PhD research focused on unsupervised representation learning of cancer tissue images by applying generative and self-supervised models.
Before starting the PhD, I worked in the semiconductor industry as a SoC Design engineer in the FPGA field at Altera Corporation and Intel Corporation in San Jose, CA. I received an M.S. in Electrical Engineering at the IIT in Chicago, and M.S. and B.S. degrees in Telecommunications Engineering at ETSIT-UPM in Madrid.
Research Experience
Publications
'Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unlabeled, unannotated pathology slides' Adalberto Claudio Quiros+, Nicolas Coudray+, Anna Yeaton, Xinyu Yang, Luis Chiriboga, Afreen Karimkhan, Navneet Narula, Harvey Pass, Andre L. Moreira, John Le Quesne*, Aristotelis Tsirigos*, and Ke Yuan*. 2024.
https://www.nature.com/articles/s41467-024-48666-7Nature Communications, 2024.
Abstract: Definitive cancer diagnosis and management depend upon the extraction of information from microscopy images by pathologists. These images contain complex information requiring time-consuming expert human interpretation that is prone to human bias. Supervised deep learning approaches have proven powerful for classification tasks, but they are inherently limited by the cost and quality of annotations used for training these models. To address this limitation of supervised methods, we developed Histomorphological Phenotype Learning (HPL), a fully unsupervised methodology that requires no expert labels or annotations and operates via the automatic discovery of discriminatory image features in small image tiles. Tiles are grouped into morphologically similar clusters which constitute a library of histomorphological phenotypes, revealing trajectories from benign to malignant tissue via inflammatory and reactive phenotypes. These clusters have distinct features which can be identified using orthogonal methods, linking histologic, molecular and clinical phenotypes. Applied to lung cancer tissues, we show that they align closely with patient survival, with histopathologically recognised tumor types and growth patterns, and with transcriptomic measures of immunophenotype. We then demonstrate that these properties are maintained in a multi-cancer study. These results show the clusters represent recurrent host responses and modes of tumor growth emerging under natural selection.
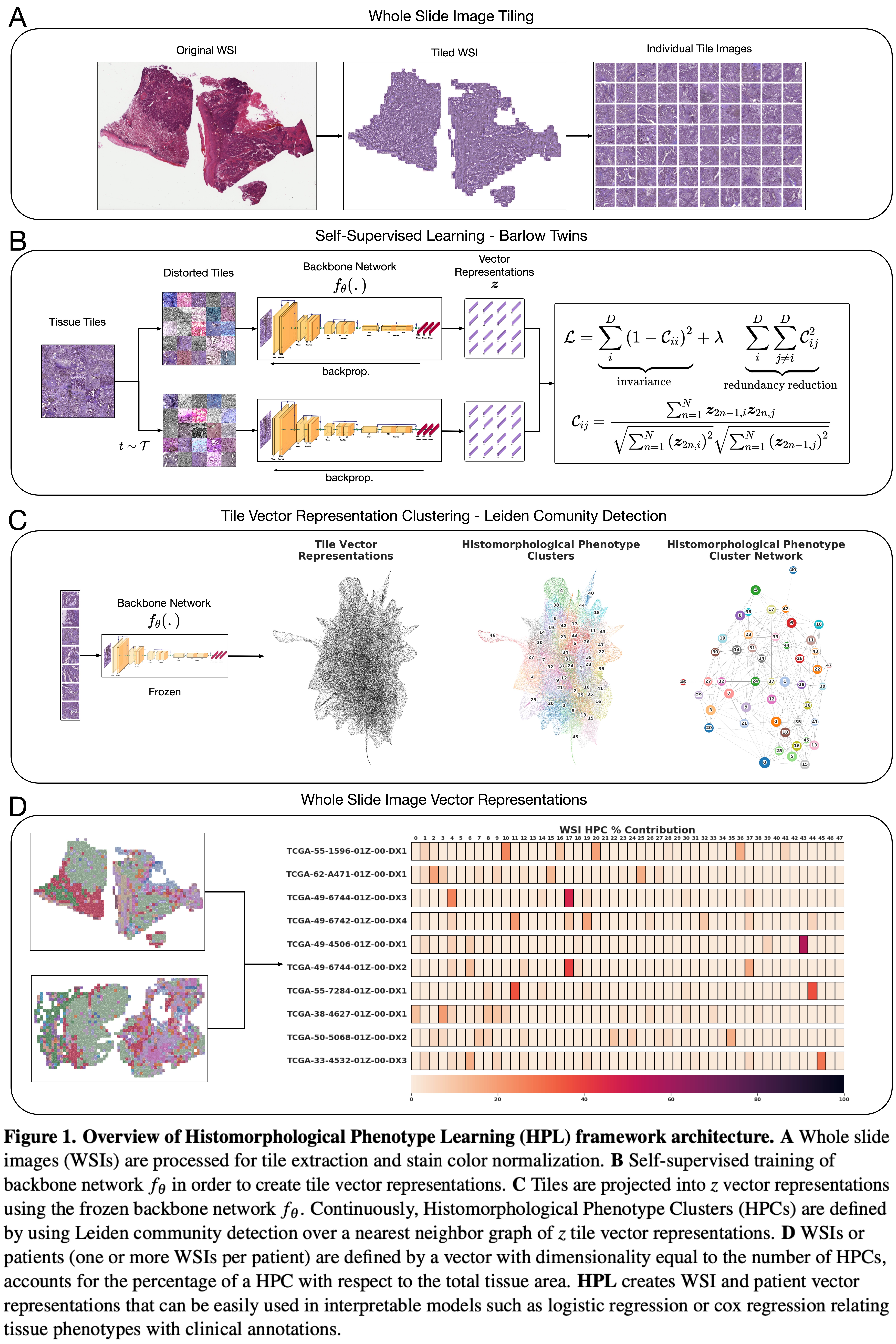
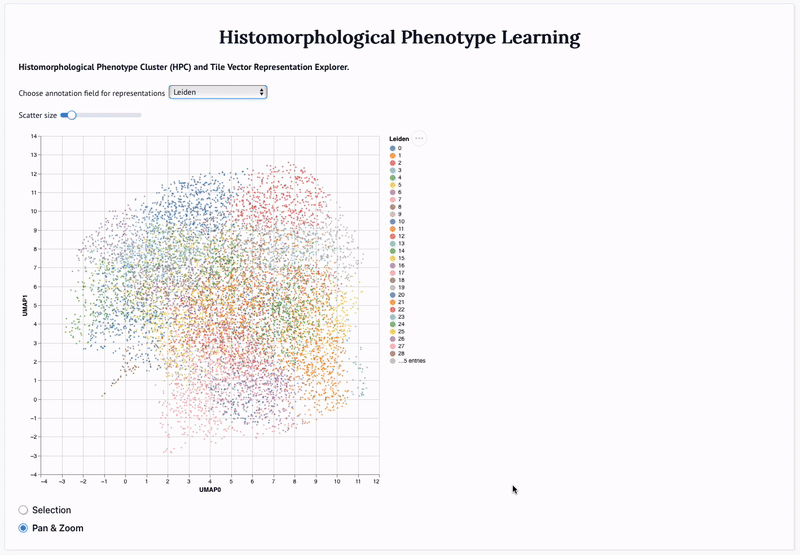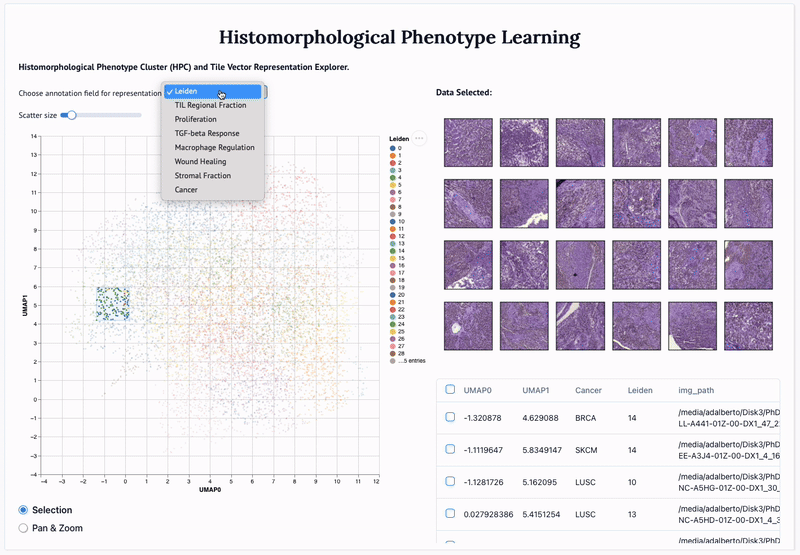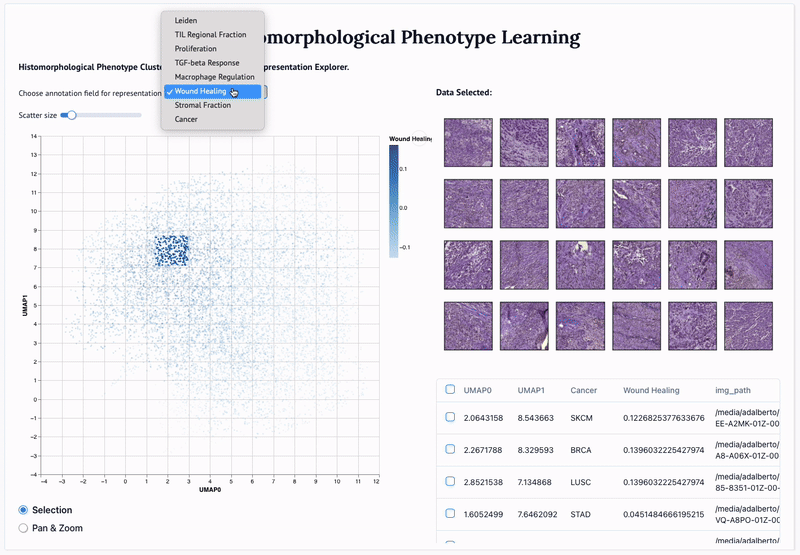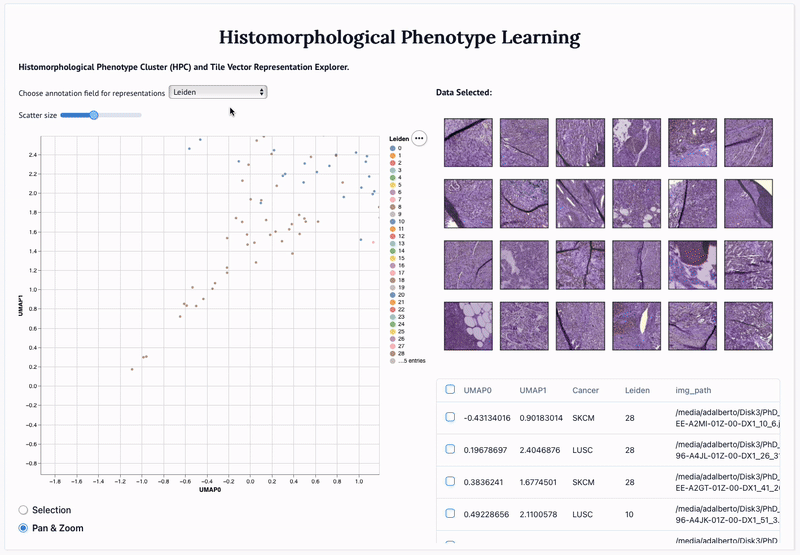
'Adversarial learning of cancer tissue representations' Adalberto Claudio Quiros, Nicolas Coudray, Anna Yeaton, Wisuwat Sunhem, Roderick Murray-Smith, Aristotelis Tsirigos, and Ke Yuan. 2021.
http://arxiv.org/abs/2108.02223Proceedings of the 24th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, 2021.
Abstract: Deep learning based analysis of histopathology images shows promise in advancing understanding of tumor progression, tumor micro-environment, and their underpinning biological processes. So far, these approaches have focused on extracting information associated with annotations. In this work, we ask how much information can be learned from the tissue architecture itself. We present an adversarial learning model to extract feature representations of cancer tissue, without the need for manual annotations. We show that these representations are able to identify a variety of morphological characteristics across three cancer types: Breast, colon, and lung. This is supported by 1) the separation of morphologic characteristics in the latent space; 2) the ability to classify tissue type with logistic regression using latent representations, with an AUC of 0.97 and 85% accuracy, comparable to supervised deep models; 3) the ability to predict the presence of tumor in Whole Slide Images (WSIs) using multiple instance learning (MIL), achieving an AUC of 0.98 and 94% accuracy. Our results show that our model captures distinct phenotypic characteristics of real tissue samples, paving the way for further understanding of tumor progression and tumor micro-environment, and ultimately refining histopathological classification for diagnosis and treatment.
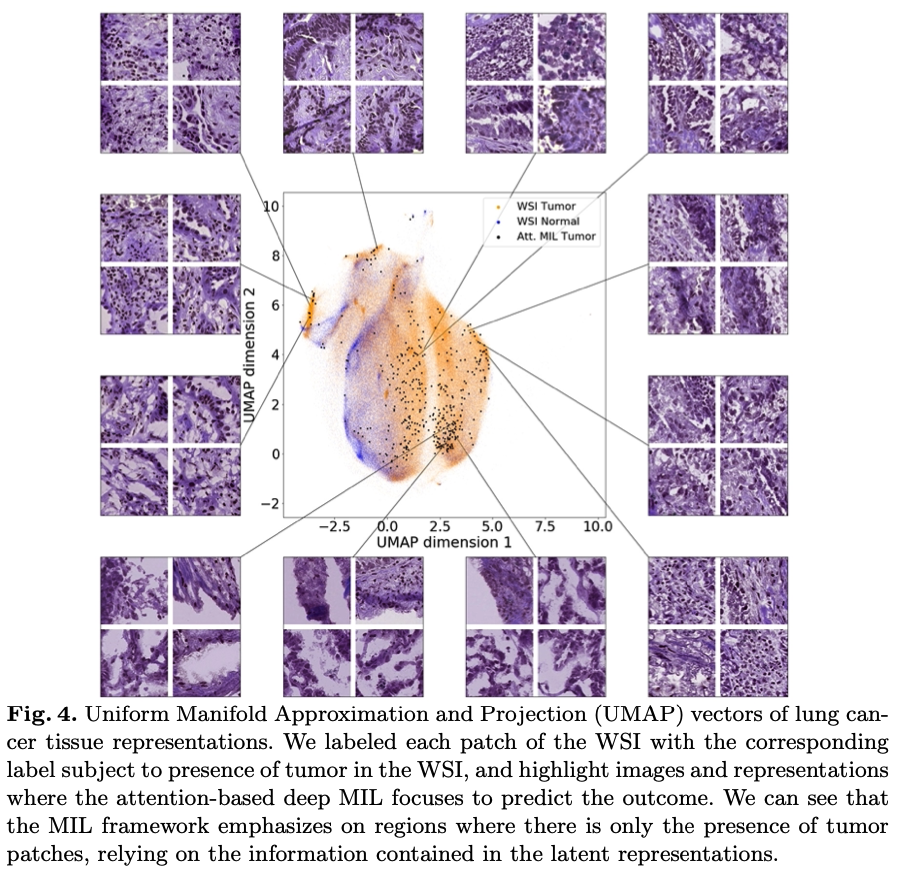
'Pathology GAN: Learning deep representations of cancer tissue' Adalberto Claudio Quiros, Roderick Murray-Smith, Ke Yuan. 2020.
https://www.melba-journal.org/article/21657Proceedings of the Third Conference on Medical Imaging with Deep Learning, PMLR, 2020.
Journal of Machine Learning for Biomedical Imaging. 2021:4. pp 1-48. Special Issue: Medical Imaging with Deep Learning (MIDL) 2020.
 Abstract: Histopathological images of tumors contain abundant information about how tumors grow and how they interact with their micro-environment.
Better understanding of tissue phenotypes in these images could reveal novel determinants of pathological processes underlying cancer, and in turn improve diagnosis and treatment options. Advances of Deep learning makes it ideal to achieve those goals, however, its application is limited by the cost of high quality labels from patients data. Unsupervised learning, in particular, deep generative models with representation learning properties provides an alternative path to further understand cancer tissue phenotypes, capturing tissue morphologies.
In this paper, we develop a framework which allows Generative Adversarial Networks (GANs) to capture key tissue features and uses these characteristics to give structure to its latent space. To this end, we trained our model on two different datasets, an H&E colorectal cancer tissue from the National Center for Tumor diseases (NCT, Germany) and an H&E breast cancer tissue from the Netherlands Cancer Institute (NKI, Netherlands) and Vancouver General Hospital (VGH, Canada). Composed of 86 slide images and 576 tissue micro-arrays (TMAs) respectively. We show that our model generates high quality images, with a Frechet Inception Distance (FID) of 16.65 (breast cancer) and 32.05 (colorectal cancer). We further assess the quality of the images with cancer tissue characteristics (e.g. count of cancer, lymphocytes, or stromal cells), using quantitative information to calculate the FID and showing consistent performance of 9.86. Additionally, the latent space of our model shows an interpretable structure and allows semantic vector operations that translate into tissue feature transformations. Furthermore, ratings from two expert pathologists found no significant difference between our generated tissue images from real ones.
Abstract: Histopathological images of tumors contain abundant information about how tumors grow and how they interact with their micro-environment.
Better understanding of tissue phenotypes in these images could reveal novel determinants of pathological processes underlying cancer, and in turn improve diagnosis and treatment options. Advances of Deep learning makes it ideal to achieve those goals, however, its application is limited by the cost of high quality labels from patients data. Unsupervised learning, in particular, deep generative models with representation learning properties provides an alternative path to further understand cancer tissue phenotypes, capturing tissue morphologies.
In this paper, we develop a framework which allows Generative Adversarial Networks (GANs) to capture key tissue features and uses these characteristics to give structure to its latent space. To this end, we trained our model on two different datasets, an H&E colorectal cancer tissue from the National Center for Tumor diseases (NCT, Germany) and an H&E breast cancer tissue from the Netherlands Cancer Institute (NKI, Netherlands) and Vancouver General Hospital (VGH, Canada). Composed of 86 slide images and 576 tissue micro-arrays (TMAs) respectively. We show that our model generates high quality images, with a Frechet Inception Distance (FID) of 16.65 (breast cancer) and 32.05 (colorectal cancer). We further assess the quality of the images with cancer tissue characteristics (e.g. count of cancer, lymphocytes, or stromal cells), using quantitative information to calculate the FID and showing consistent performance of 9.86. Additionally, the latent space of our model shows an interpretable structure and allows semantic vector operations that translate into tissue feature transformations. Furthermore, ratings from two expert pathologists found no significant difference between our generated tissue images from real ones.
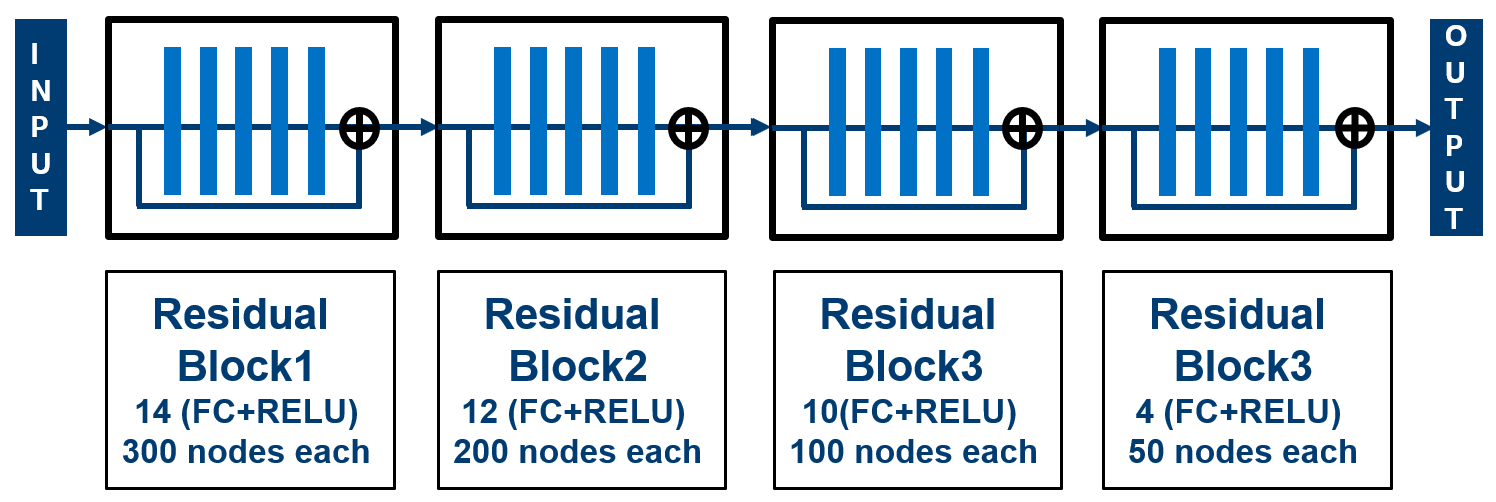
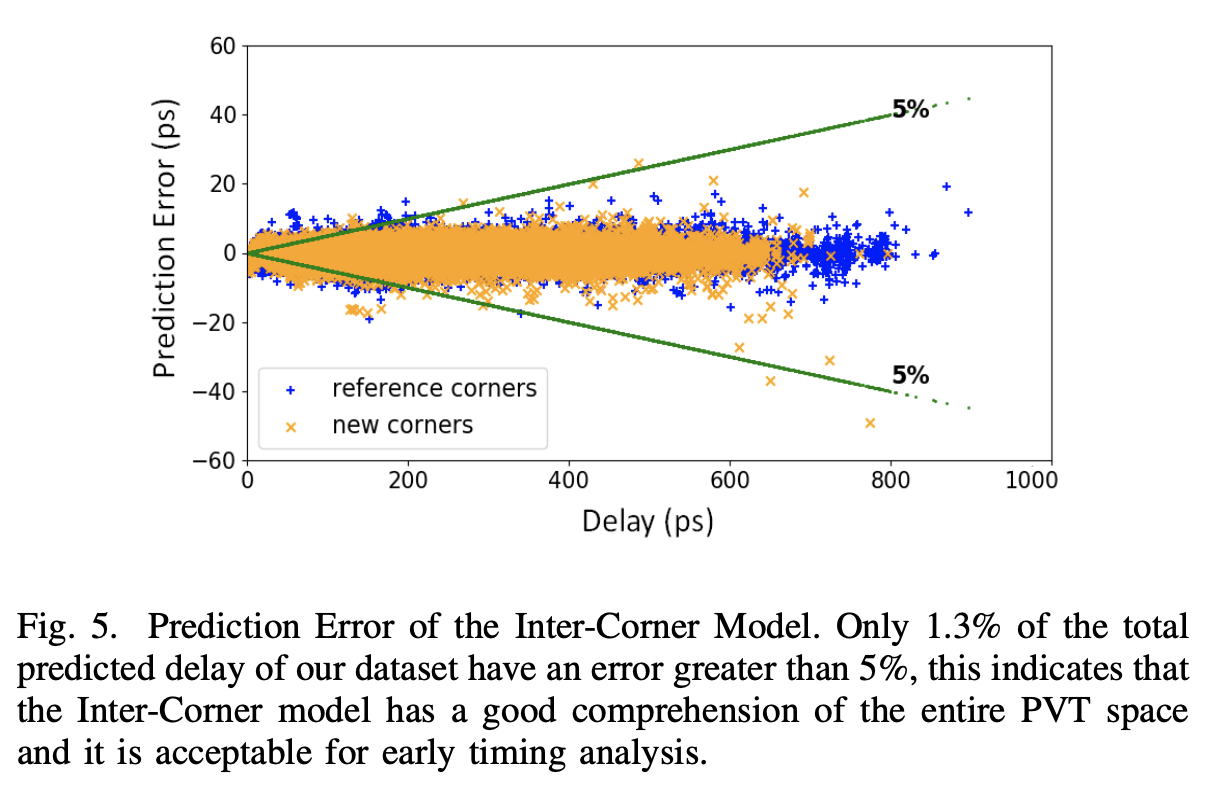
'DNNLibGen : Deep Neural Network Based Fast Library Generator' Eunice Naswali, Adalberto Claudio Quiros, Pravin Chandran. 2019.
https://ieeexplore.ieee.org/document/896519126th IEEE International Conference on Electronics Circuits and Systems.
 Abstract: We propose a new modeling methodology using deep learning techniques for generating timing
models for Static Timing Analysis (STA). Current device behavior is non-linear, non-monotonic and exhibits high sensitivity to (Process Voltage Temperature) PVT variation which imposes a
myriad of design challenges including the need for analysis at several PVT corners. While complete PVT coverage is crucial for detecting design issues early and achieving time-to-market goals
with improved predictability, the number of PVT corners are growing exponentially and library generation has also become a significant bottleneck in current design cycles. To this end, we have
developed a novel methodology for timing library generation that uses data from sparse characterization in PVT space and generates delay models at required sign-off corners.
We have employed deep neural nets with residual connections for delay modeling and our methodology enables a ‘single model’ to fully comprehend multiple cell types, PVT corners
and generate required PVT timing libraries. The proposed library-generator uses a novel inter-corner model to generate delay tables at 17 test corners using 7 corners as reference.
In addition, we have developed an intra-corner model, to generate dense 8x8 delay tables using delays from 10 slew/load points as reference. The results show that, using these models,
we are able to achieve key improvements with over 98.7% of calculated delays within acceptable tolerance while reducing characterization run-time for early milestones by up to 60%.
Abstract: We propose a new modeling methodology using deep learning techniques for generating timing
models for Static Timing Analysis (STA). Current device behavior is non-linear, non-monotonic and exhibits high sensitivity to (Process Voltage Temperature) PVT variation which imposes a
myriad of design challenges including the need for analysis at several PVT corners. While complete PVT coverage is crucial for detecting design issues early and achieving time-to-market goals
with improved predictability, the number of PVT corners are growing exponentially and library generation has also become a significant bottleneck in current design cycles. To this end, we have
developed a novel methodology for timing library generation that uses data from sparse characterization in PVT space and generates delay models at required sign-off corners.
We have employed deep neural nets with residual connections for delay modeling and our methodology enables a ‘single model’ to fully comprehend multiple cell types, PVT corners
and generate required PVT timing libraries. The proposed library-generator uses a novel inter-corner model to generate delay tables at 17 test corners using 7 corners as reference.
In addition, we have developed an intra-corner model, to generate dense 8x8 delay tables using delays from 10 slew/load points as reference. The results show that, using these models,
we are able to achieve key improvements with over 98.7% of calculated delays within acceptable tolerance while reducing characterization run-time for early milestones by up to 60%.
Conference Workshops
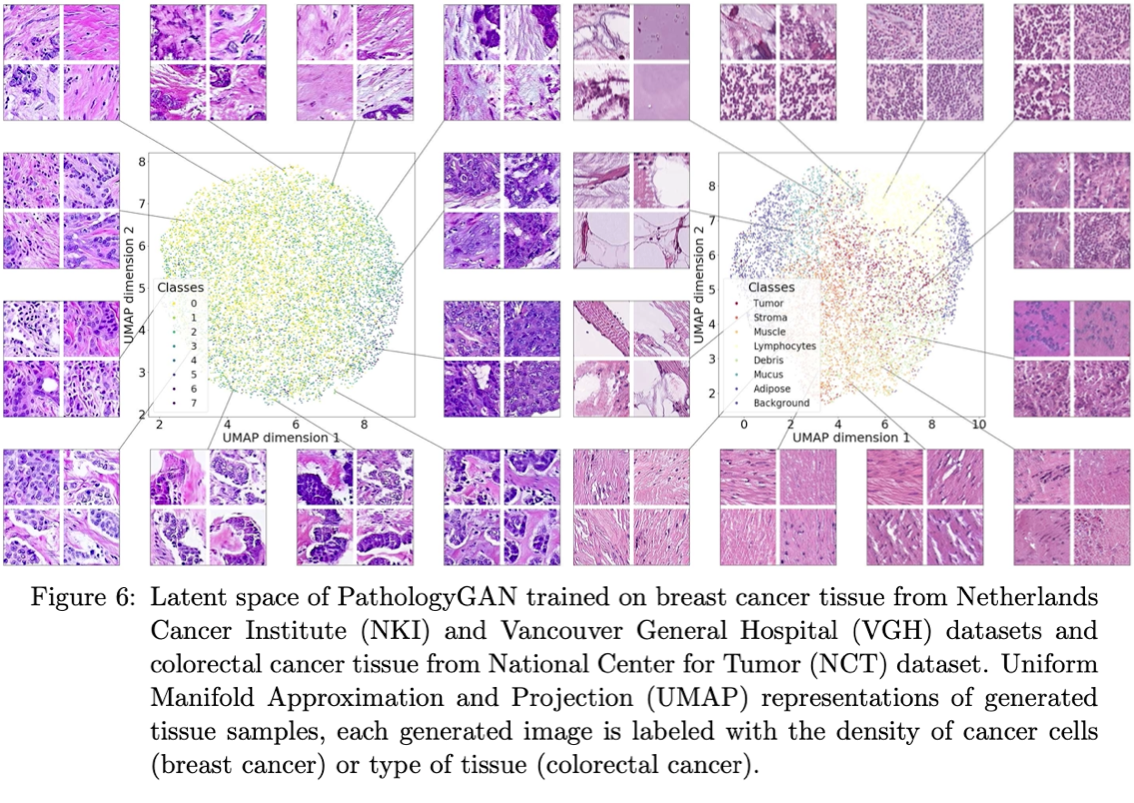
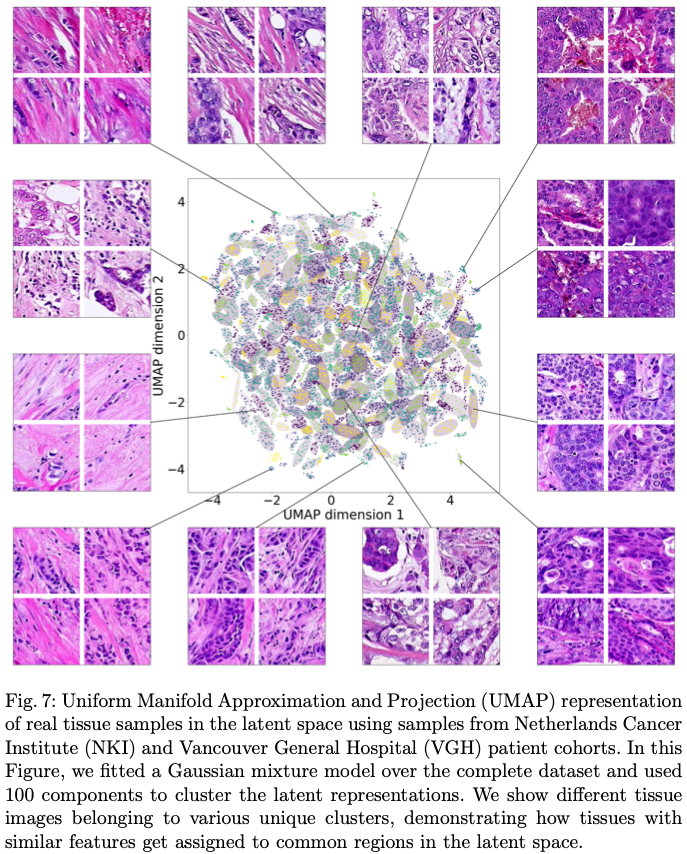
'Learning a low dimensional manifold of real cancer tissue with Pathology GAN' Adalberto Claudio Quiros, Roderick Murray-Smith, Ke Yuan. 2020.
http://arxiv.org/abs/2004.06517NeurIPS 2020 Learning Meaningful Representations of Life Workshop.
Late breaking research talk at RECOMB 2020 Computational Cancer Biology.
Abstract: Application of deep learning in digital pathology shows promise on improving disease diagnosis and understanding. We present a deep generative model that learns to simulate high-fidelity cancer tissue images while mapping the real images onto an interpretable low dimensional latent space. The key to the model is an encoder trained by a previously developed generative adversarial network, PathologyGAN. We study the latent space using 249K images from two breast cancer cohorts. We find that the latent space encodes morphological characteristics of tissues (e.g. patterns of cancer, lymphocytes, and stromal cells). In addition, the latent space reveals distinctly enriched clusters of tissue architectures in the high-risk patient group.
Miscellaneous
- March, 2023: Transformers’ Tutorial Slides: Attention & Transformers, Training & Behavior, NLP & Modifications, Vision Transformers.
- May, 2022: Gave a talk at the MELBA symposium on PathologyGAN: Slides.
- June - September, 2021: Worked as an AI research intern at Arabesque AI on time-series GANs.
- June, 2020: Gave a talk at late breaking research RECOMB-Computational Cancer Biology 2020 on ‘Learning a low dimensional manifold of real cancer tissue with PathologyGAN’: Slides
- March, 2020: Gave a lecture at the ‘Deep Learning’ course of MSc in Data Science, introducing Generative Adversarial Networks (GANs): Slides
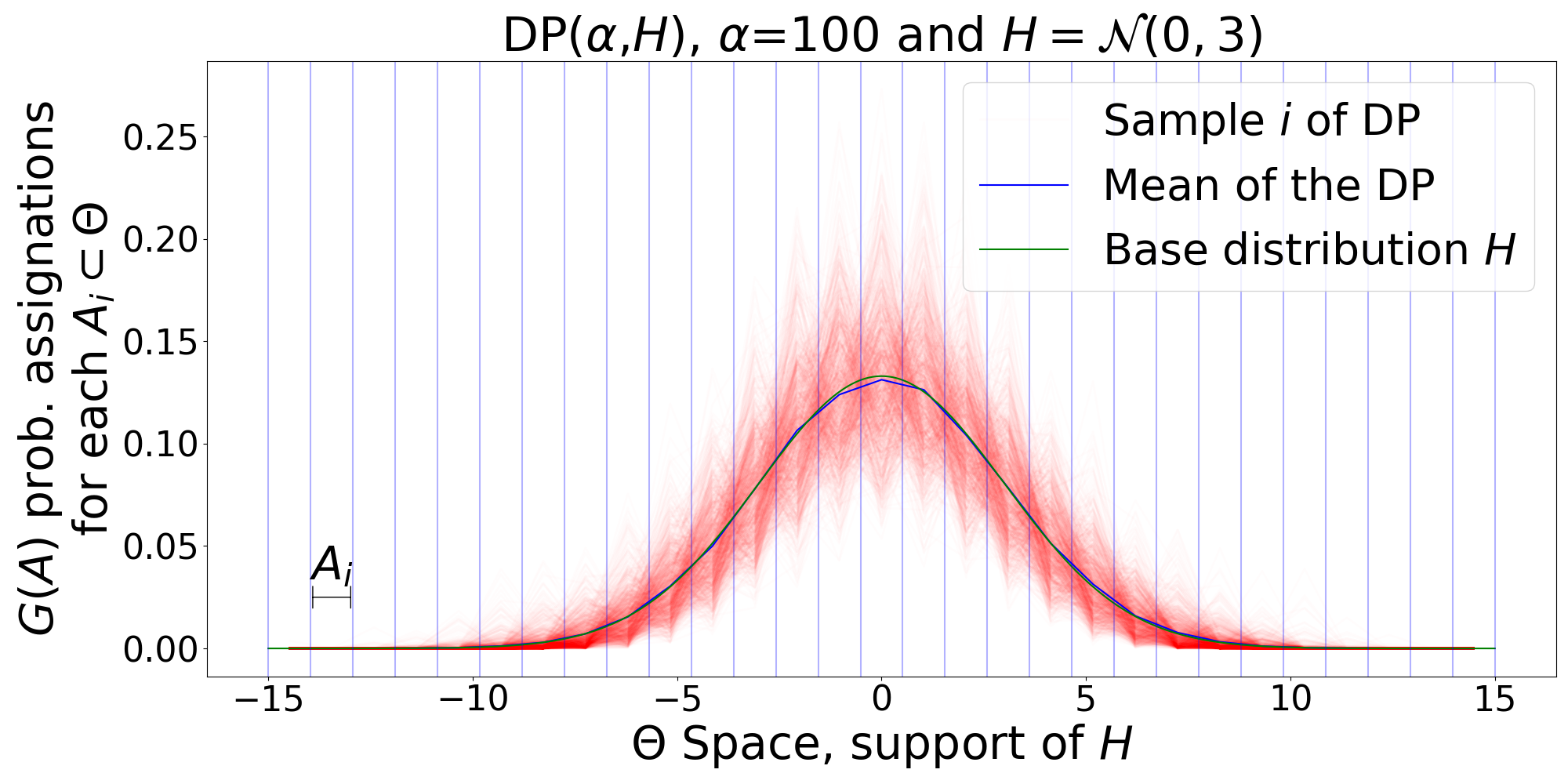
- January, 2020: Wrote a tutorial document and code implementations of related concepts to Dirichlet Processes: GEM distribution, Polya Urn, Chinese restaurant process, Stick-Breaking construction, DP Posterior, and DP Mixture Models.
- October - December, 2020: Worked as a teaching assistant and gave two lectures at the ‘Machine Learning for Data Scientists’ course of MSc in Data Science, briefly introducing Sampling methods and Variational Inference: Sampling slides, MCMC Bayesian Linear regression example, Variational Inference slides, and VI Bayesian Linear regression example
- March, 2019: Wrote a brief survey on GANs: slides and code for relevant models.
Professional Experience
Intel Corporation
San Jose, CA, USASenior SoC Design Engineer - Fabric Performance Leadership Team
April 2017 – September 2018
I worked in the Fabric Performance Leadership team, our goal was to analyze the Stratix 10 FPGA design from the hardware and software perspective to find flaws and push the FPGA frequency performance forward.
During this period, a few of the most important achievements were:
- A Python graph builder tool that generates a graph of the FPGA’s routing structure, prunes it and creates a visualization from an register-transfer level netlist design.
- Implementated a device End-of-Life (EOL) methodology tool that obtained the aging degradation delay on FPGA’s transistors.
- Analyzed logical and physical implementation of Adders: From the FPGA software, integrated circuit design and architecture perspective. The result of this study was a redesign of the adder’s implementation leading to a 70% improvement on frequency performance for adders.
- Published ‘DNNLibGen : Deep Neural Network Based Fast Library Generator’ Eunice Naswali, Adalberto Claudio Quiros, Pravin Chandran.
Altera Coporation/Intel Corporation
San Jose, CA, USASenior SoC Design Engineer - Full Chip Timing Team
June 2014 – April 2017
Worked in the Full Chip Timing team on Static Timing Analysis, first to verify the hardware FPGA designs running at the frequency specifications, and secondly to correlate the FPGA software models with the hardware designs and silicon devices.
- Developed a Stratix 10 full chip timing violation tool: This tool gathers all full chip timing violations due to maximum transition (~100K instances), cross reference each violation to a lower system blocks, and compiles them for each block designer.
- Arria 10 Frequency binning and register to register timing correlation lead: Silicon/Quartus-FPGA-SW/HSPICE model frequency correlation, given an internal Altera Quality Award in 2015 Q2.
- Developed a timing tool in Python, HSPICE, and Quartus FPGA Software significantly impacting the process: Increasing the number of data points from 10 to 7K, with different features including Power-Voltage-Temperature (PVT), voltage threshold and sheet resistance sweeping options.
Worked developing bots for web data scraping, additionally I help in maintaining and developing improvements on middle tier code and servers, which controled the bots and jobs executions. Most of this work was done in SQL, C#, Python, C++.
Education
University of Glasgow
Doctor of Philosophy in Computing Science, Machine Learning
Glasgow, Scotland, U.K.
Illinois Institute of Technology
Master of Science in Electrical Engineering
Chicago, IL, USA
Polytechnic University of Madrid, ESTIT-UPM
Master & Bachelor of Science in Telecommunications Engineering
Madrid, Spain